Introduction
In fact, nature contains a huge diversity of bioactive molecules which have therapeutic activity. Utilization of natural anticancer agents has great potential to overcome cancer and yield promising results [1]. Venomous animals play a vital role in the discovery of novel therapeutic candidates. They provide various venoms to prevent themselves from external surroundings [2]. The venom gland of snakes has been the subject of numerous studies due to the highly anticancer action of their secretions and the evolutionary significance of this specialized trophic and defensive adaptation. Scientists over the past few decades have postulated that despite of the harmful and life-threatening effects of snake venom, its component may provide highly specific research tools for the development of novel life-saving medicines and drugs against some common and life-threatening diseases. Egypt is a rich source of flora and fauna is also home to a rich diversity of snake's species. However, few studies are reported about the venom gland from Egypt [3-6]. Shanbhag et al. (2015) [7] explored the potential of snake venoms in treating cancer. They noted that snake venoms have been used as traditional medicine for centuries and their components have a high specificity and efficacy in targeting cells and their components. Furthermore, snake venoms exhibit greater cytotoxicity towards tumor cells compared to normal cells, and have several therapeutic actions that make them an attractive option for cancer treatment. Recent advancements in technology have enabled the identification and extraction of new therapeutic components from snake venoms in a shorter time frame.
The venoms from various snakes are important agents for the treatment of several types of cancers. As stated previously in the literature cites, several toxins from snake venom have shown promise as potential candidates for future ovarian cancer treatments [8], hepatocellular carcinoma (HCC) [9, 10], colorectal cancer [11], breast cancer (MCF-7) cells [12-17], colon cancer [15, 18, 19], human cervical cancer cells (HeLa) [20], and fibroblast cells (HFF) [20, 21]. Locally, Salama et al. (2018) [22] conducted a study on the L-amino acid oxidase enzyme (Cv-LAAOI) isolated from the venom of the Egyptian Sand viper (C. vipera). against the breast cancer cell line (MCF-7), with an IC50 value of 2.75 ± 0.38 μg/ml, compared to different tumor cell lines (liver HepG2, lung A549, colon HCT116, and prostate PC3). Also, Abdelglil et al. (2021) [23] conducted a study on the potential anticancer properties of crude and irradiated C. cerastes snake venom, as well as propolis ethanolic extract. The researchers found that the irradiated venom showed higher toxicity than crude venom, while the combination of crude venom and propolis extract demonstrated the highest toxicity against lung and prostate cancer cells. Sadat et al. (2023) [19] investigated the potential of Oxineur, a novel peptide from Caspian cobra N. najaoxiana against colon cancer. In vivo assay, Mahmoud et al. (2023) [24] conducted a study to assess the effect of C. cerastes LAAO on some hematological parameters in hepatocellular carcinoma-induced in the rats. Moreover, screening of wide range of venoms (61 taxa) from the major venomous snake families (Viperidae, Elapidae, and Colubridae) to investigate their anticancer effects on MCF-7 breast cancer cells and A-375 melanoma cells was investigated. 3-(4, 5-dimethylthiazol-2-yl)-2, 5- diphenyltetrazolium bromide (MTT) cell viability assays showed that most of the venoms had significant anticancer effects [14]. So, the study aims to make compare anticancer effects of some snakes from the Egyptian environment through cytotoxicity on two types of carcinomic cells (in vitro assay), biomarkers profile, and DNA adducts.
Materials and Methods
The experiment adhered to the guidelines of the ethical committee of Tanta University, Tanta, Egypt. We obtained four mature specimens from five Egyptian venomous snakes' species that differ in their habitats and are healthy. The first one is Egyptian Cobra, Naja haje (N.H) which was obtained from Alwastaa district, Beni Suef Governorate. The second one is painted saw-scaled viper, Echis coloratus (E.C) which was obtained from Saint Catherine district, South Sinai Governorate. The third one was Egyptian saw-scaled viper, Echis pyramidum (E.P) obtained from Fayoum desert, Fayoum Governorate. The fourth one is Sahara Sand Viper, Cerastes vipera (C.V) which was obtained from Alhizam al'akhdar, Giza Governorate. The last one is Saharan Horned Viper, Cerastes cerastes (C.C) which was obtained from Fayoum desert, Fayoum Governorate. Animals were kept in plastic cages in a room under standard condition of illumination with a 12-hr light–dark cycle at 28 °C. They were provided with water and balanced diet. All samples were collected between August and October 2021.
Venom collection and preparation
The snake venom was collected by milking, where the snakes were allowed to bite onto a parafilm-sealed sterile collection cup, and then stored at −80 °C until used. The collected venoms were put in Falcone tubes (45 ml) and subjected to freeze-drying process. The drying program was done at -60 °C for 22 hrs and vacuum level (10-3 mbar). The remaining powders were stored at -80 °C until used.
Cell Lines
Cell Culture
Two types of the cells: breast cancer cells (MCF-7) and hepato-cancer cells (HepG2) were proved by VACCIRA, Giza, Egypt. The cells were maintained in a standard medium consisting of DEMEM with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin and incubated at 37 °C and 5% CO2 prior to use. The media were replaced with fresh DEMEM-10% in phosphate-buffered saline (PBS). The cells were maintained by sub-culturing them after arriving at an acceptable confluence.
Anticancer potency
The cells were seeded into 96-well cell culture plates at a concentration of 1 × 104 cells/ml and incubated for 24 hr under standard conditions to reach exponential growth. The cells were treated with different concentrations of the examined toxins ranged from 0.025 to 0.4 μg/ml. After 24 hr of incubation, the medium was removed and 5 mg/ml of 3-(4, 5- dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) reagent was added to each plate and left to incubate for 3-4 hrs. The formazan crystals were dissolved in 100 μl acidified isopropanol and read at 630 nm by using an ELISA microplate reader (Bio-RAD microplate reader, Japan). Each concentration was repeated in triplicates. The fields of untreated and pesticide-treated cells were visualized on light microscope to compare the defects on the cells after the exposure period. The Medium lethal inhibition (IC50) values were calculated by using Excel software program and probit Lpd line program. Cell viability was calculated as follows:
Where, AbsS and AbsC were absorbance of the cells incubated with toxin and free toxin, respectively.
Biomarkers for cytotoxic effects
Two levels: 1/10 and 1/50 IC50, for each toxin were used. For each concentration, 3 replicates were used in a T-25 culture flask as described above. After 24 hr of the incubation, the medium was removed and the remaining cells were harvested in phosphate buffered saline (PBS) and centrifuged at 2000 rpm for 10 min at 4 °C. For enzyme assays, the media and cell lysate were stored at −80 °C until used. Cell Lysate (pellets) were homogenized in cold (50 mM potassium phosphate buffer pH 7.5, and 2.0 mM EDTA). The homogenate was centrifuged at 4000 rpm and 4 °C for 15 min. The homogenate was used as a source for lactate dehydrogenase (LDH), lipid peroxidation (LPO), and reduced glutathione (GSH) content assays.
Lactate dehydrogenase (LDH)
The enzyme activity was measured by the method of Mc Queen (1975) [25] by using sodium pyruvate as a substrate. The activity was expressed as U/L.
Lipid peroxidation (LPO)
Thiobarbituric acid reactive substances (TBARS) were used as an index of LPO according to Rice-Evans et al. (1991) [26]. TBARS was determined by spectrophotometric quantification of the malondialdehyde (MDA) content. An aliquot (250 μl) of cell lysate or media was mixed with 1 ml of 15% (w/v) trichloroacetic acid (TCA) in 25 mM HCl and 2 ml of 0.37% (w/v) thiobarbituric acid (TBA). The mixture was boiled for 10 min, quickly cooled, and immediately centrifuged at 5000 rpm for 5 min. The absorbance was determined at 535 nm. MDA was quantified using an extinction coefficient of 156 mM1, and its concentration was expressed as mM/g tissue.
Glutathione (GSH) content
The method was based on the reduction of 5, 5′-dithiobis 2-nitrobenzoic acid (DTNB) with GSH to produce a yellow compound measured at 405 nm [27]. To 500 μl of enzyme source, the same volume of 500 mM TCA was added, followed by centrifugation at 3000 rpm for 15 min. An aliquot (500 μl) was well mixed with 1 ml of each 100 mM PBS buffer, pH 7.4, and 1 mM DTNB. After 10 min, the absorbance was measured at 405 nm against the blank. The GSH concentration was expressed as nM/g tissue.
Genomic diversity
Extraction and purification of DNA
Suspensions of the two types of cultured mammalian cells were centrifuged at 250 xg for 5 min. The supernatant was discarded and re-centrifuged again. The pellet was rinsed with PBS to remove residual medium. The sample was suspended in 200 μl, followed by the same volume of lysis solution and 20 μl of Proteinase K solution and mixed thoroughly by vortex. The mixture was incubated at 56 C with shaking for 10 min. Then, 20 μl of RNase solution was added, mixed and incubated for 10 min at room temperature. An aliquot (400 μl) of 50% ethanol was added and mixed again. The prepared lysate was transferred to Gene JET Genomic DNA Purification Column inserted in a collection tube (Genomic DNA purification #K0721 #0722, Pub. No.MAN0012663). Then, it was centrifuged at 6000 xg for 1 min. The extract was discarded and the purification Column inserted into a new 2 ml collection tube.
An aliquot (500 μl) of wash buffer I (with ethanol) was added to the purified column, centrifuged at 8000 xg for 1 min. The extract was discarded and the column was placed back into the collection tube. The same volume of wash buffer II (with ethanol) was added and centrifuged again for 3 min at maximum speed (≥12000 xg). The extract was discarding again and the column was transferred to a sterile 1.5 ml micro centrifuge tube (not included). An aliquot (200 μl) of elution buffer was added, incubated for 2 min at room temperature and centrifuged for 1 min at 8000 xg. The purification column was discarded, and the purified DNA was stored at -20 °C immediately.
The purify factor (A260/A280 nm) was quantified spectrophotometrically to obtain the ratio between genome/protein value.
ISSR conditions and preparation
The PCR amplification was performed using three simple sequences repeats (ISSRs) primers to differentiate and fingerprint the samples of cultured mammalian cells (HepG2 and MCF-7) treated with five Egyptian snake venoms extracts. A total volume (25 µ l) was used for the amplification as follows: 2 µ l DNA, 2 µ l primer, 2.5 µ l 10X buffer, 2.5 µ l MgCl2 (50 mM), 2.5 µ l dNTPs (4 mM), and 0.2 µ l (5 mM/µ l) Taq-polymerase.
The procedures were carried out in automated thermal cycler with the following PCR condition: one cycle at 94 °C for 10 min, followed by 35 cycle (1 min at 94 °C, 1 min of annealing for primer at 32 °C, and 1 min at 72 °C and final temperature was done at 72 °C. Gel electrophoresis in an agarose gel 2% in TAE buffer with 105 v for 40 min was used to separate PCR products. The bands were then noticed with ethidium bromide (EtBr) staining and photographed with a gel documented system (Bio-Rad Gel Doc., 2000).
Results
Anticancer potency
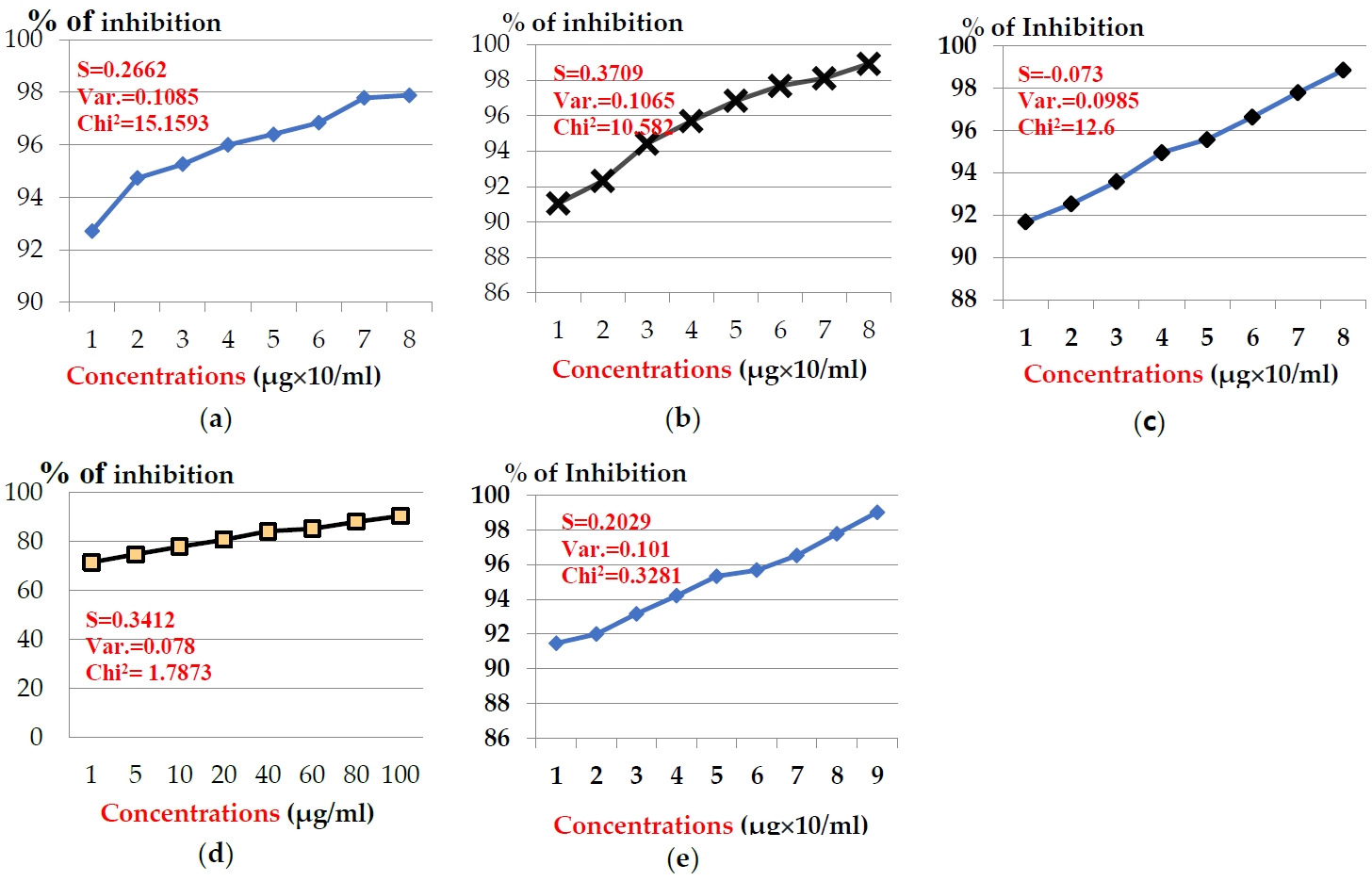
The anticancer effects of venoms extracted from five Egyptian snakes on hepato-carcinomic cells (HepG2) and breast cancer cells (MCF-7) was assayed using MTT assay, which revealed that venom’s anticancer potency was estimated and cell-type-dependent, as viability increased as long as concentration decreased. The presented data showed that, the examined venoms were more potent on MCF-7 cells than HepG2 cells. In case of MCF-7, the examined toxins showed significant inhibition in cell generation after the incubation time (24 hr) (Figure 1). Concentrations of snake venom, E. pyramidum (E.P) in the range from 1 to 100 μg/ml exhibited inhibition % from 92.73 to 97.89 % after the incubation period (24 hr) (Figure 1a). Snake venom, C. vipera (C.V) in the same concentrations exhibited inhibition % (91.05 - 98.91 %) (Figure 1b). The same pattern was obtained for snake venom, N. haje (N.H), where the same concentrations exhibited inhibition % range (91.68 - 98.84%) (Figure 1c). Slight inhibition % was noticed for the concentrations of snake venom, E. coloratus (E.C), where they exhibited inhibition % in the range 71.47 - 90.52 % (Figure 1d). Finally, snake venom, C. cerastes (C.C) at concentrations: 0.5-100 μg/ml exhibited inhibition % ranged from 91.47 to 99.02 % (Figure 1e). The examined toxins exhibited the medium inhibition concentration (IC50) values: 3.48, 3.60, 3.70, 4.33, and 4.49 μg/ml for venoms: E.P, C.V, N.H., E.C, and C.C, respectively.
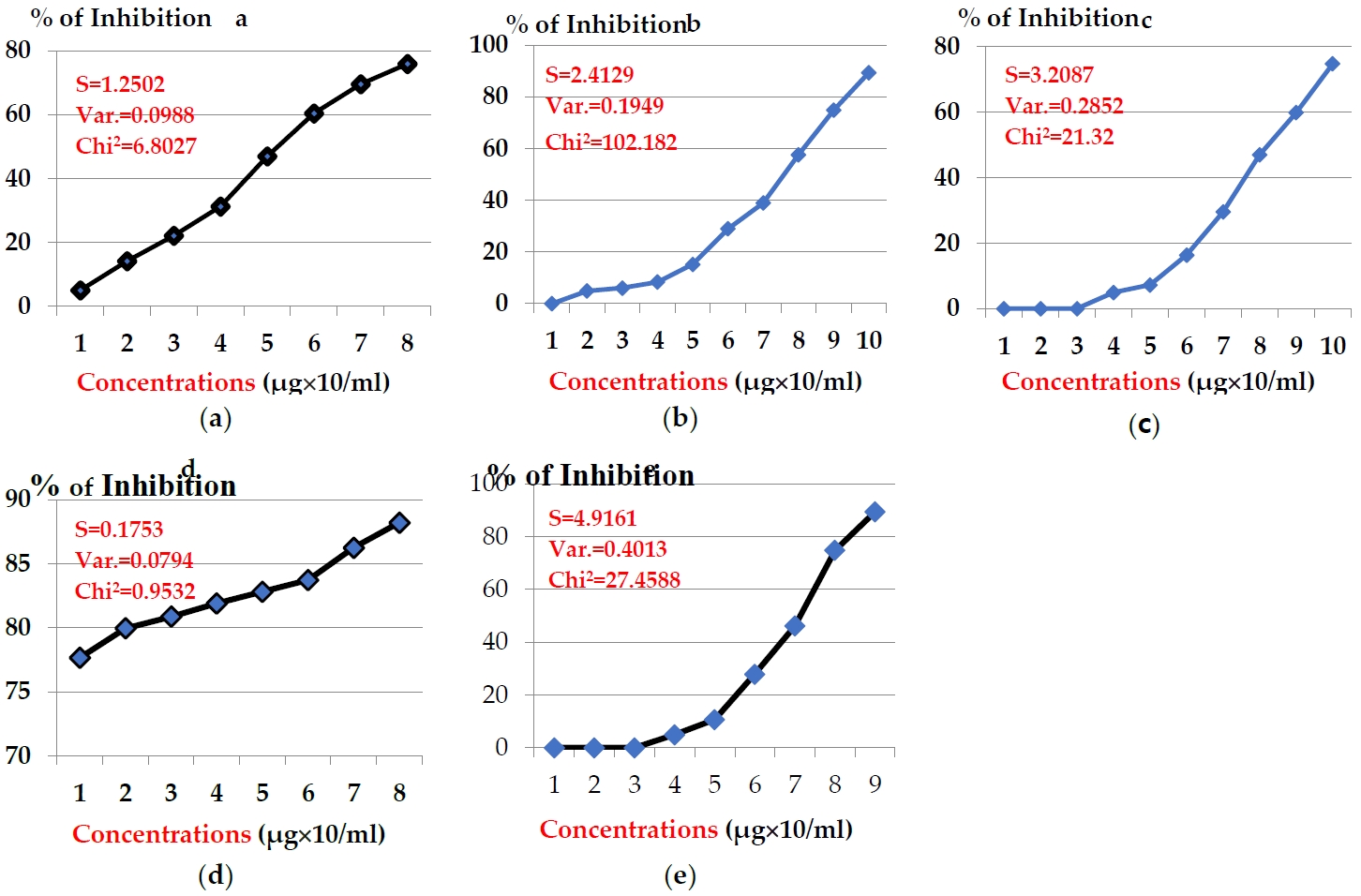
The anticancer potency of the above toxins in hepato-carcinomic cells (HepG2) showed another pattern, indicating efficiency less than MCF-7 as illustrated in Figures 2. The concentrations of snake venom E.C. showed inhibition % in the range 77.66 - 88.21 % (Fgure 2a). Potency of snake venom E.P exhibited slight % of inhibition in range of 4.93 - 75.95 % (Figure 2b). While, the potency of snake venom C.V. exhibited another range of inhibition % (4.93 - 89.35 %) (Figure 2c). Significant decline in the potency of snake venom C.C. was noticed, where its concentrations exhibited the inhibition % range from zero to 89.35% (Figure 2d). Also, significant decline was noticed in anticancer potency of snake venom N.H., where the concentrations (20-140 μg/ml) exhibited inhibition % in the range (4.93 - 74.80 %) (Figure 2e). The values of IC50 after 24 hr were 4.32, 17.77, 59.72, 63.75, and 217.90 μg/ml for toxins: E.C, E.P, C.V, C.C, and N.H, respectively.
Biomarkers for cytotoxic effects
Lactate dehydrogenase (LDH)
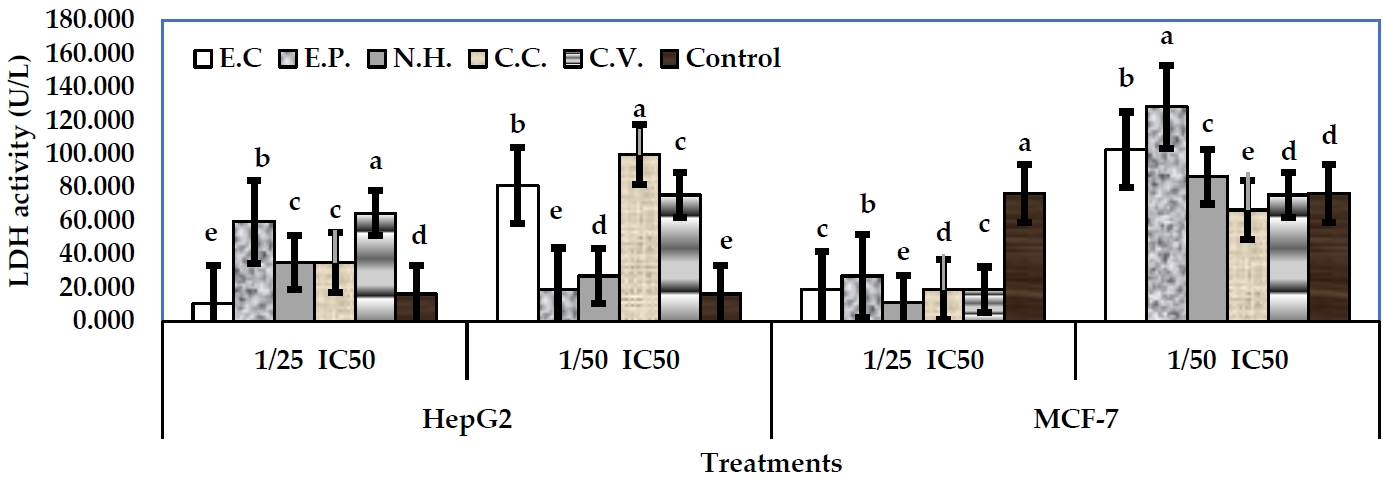
The activity of LDH was conducted as a biomarker for cytotoxicity assessing. The examined toxins from snake venom showed significant alterations in LDH activity in the treated cells in media or cell lysate, respect to their controls. In case of MCF-7 cells, sub-lethal concentrations: 1/25 and 1/50 IC50 were examined. In fact, 1/25 IC50 levels were more effective than 1/50 IC50 levels. In well media, 1/25 IC50 level of N.H. exhibited the greatest decline in LDH activity (11.13 U/L) (P<0.05), followed by E.C, C.C, and C.V. (18.89 U/L), respect to their control (76.28 U/L) (Figure 3). However, 1/50 IC50 levels of all toxins significantly exhibited increases in enzyme activity, respect to their control. Regarding cell lysate, 1/25 IC50 level of E.P exhibited the greatest decline in enzyme activity (16.19 U/L) (P<0.05), followed by C.V (18.89 U/L) (P<0.05), N.H (35.08 U/L), E.C (39.28 U/L), and C.C (59.36 U/L), respect to their control (75.36 U/L) (Figure 4). Level 1/50 IC50 of C.V exhibited the greatest decline in the enzyme activity (10.79 U/L) (P<0.05), followed by N.H (51.27 U/L). Other toxins exhibited significant increases in LDH arising the values: 97.14, 72.85, and 70.16 U/L for E.C, E.P and C.C, respectively.
In case of HepG2 cells, 1/25 IC50 of E.C exhibited the greatest decline of LDH activity in media (10.79 U/L) (P<0.05), respect to control (16.19 U/L). Other toxins at the same level significantly exhibited increases in the enzyme activity arising the values: 59.36, 35.08, 35.08, and 64.47 U/L for E.P, N.H, C.C and C.V, respectively (Figure 3). All toxins at 1/50 IC50 levels exhibited significant increases in LDH activity arising the values: 80.95, 18.89, 26.98, 99.84, and 75.55 U/L for E.C, E.P, N.H, C.C and C.V, respectively. Regarding cell lysate, 1/25 IC50 levels of the examined toxins induced significant declines in LDH activity with values: 24.29, 40.48, 48.57, 26.55, and 32.38 U/L for E.C, E.P, N.H., C.C, and C.V, respectively, respect to control (73.60 U/L) (Figure 4). On the other hand, 1/50 IC50 level of C.V exhibited the greatest decline in LDH activity (18.89 U/L), followed by E.C (24.29 U/L), and C.C (40.48 U/L), respect to control. Other toxins exhibited significant increases of the activity (E.P, 639.51 U/L, and N.H, 59.36 U/L).
Malondialdhyde (MDA)
Lipid peroxidation (LPD) profile was assessed through determination of MDA concentration in the cell lysate. MDA is used as biomarker of oxidative stress of xenobiotic in the cellular membranes. So, damage of the cells can be diagnosed by using such indication. In MCF-7 cells, level 1/25 IC50 of toxins exhibited declines in MDA content, respect to their control (Figure 5). In MCF-7 cells, 1/25 IC50 level of venom C.C exhibited the greatest decline in MDA levels (0.33 nM/g tissue), followed by E.C. (0.39 nM/g tissue), and C.V (0.40 nM/g tissue). Slight decline in MDA (0.43 nM/g tissue) was induced by E.P treatment, respect to control (0.50 nM/g tissue). Level of 1/50 IC50 of all toxins exhibited increases in MDA values, respect to their control.
Regarding HepG2 cells, 1/25 IC50 levels of all toxins exhibited significant declines in MDA levels arising the values: 0.33, 0.26, 0.33, 0.31, and 0.34 nM/g tissue (P<0.05) for toxins: E.C, E.P, N.H, C.C and C.V, respectively, respect to control (0.51 nM/g tissue). In case of 1/50 IC50, venom N.H exhibited significant decline in MDA level (0.34 nM/g tissue), followed by C.C (0.46 nM/g tissue). However, other toxins exhibited significant increases in MDA levels arising the values: 1.35, 1.37, and 1.32 nM/g tissue (P<0.05) for toxins: E.C, E.P, and C.V, respectively (Figure 5).
Glutathione (GSH)
Such this biomolecule is an essential for cell defense. So, it is assayed as biomarker for oxidative stress induction in the cells. In case of MCF-7 cells, 1/25 IC50 levels of all toxins exhibited non-significant increases in GSH content (0.084, 0.079, 0.083, 0.325, and 0.082 U/mg protein), for toxins: E.C, E.P, N.H, C.C and C.V, respectively, respect to their control (0.083 U/mg proteins). Level 1/50 IC50 of E.P showed significant decline in GSH content (0.073 U/mg protein), followed by C.C (0.076 U/mg protein). Other toxins exhibited increases in GSH content, respect to their control (untreated) (Figure 6). Regarding HepG2 cells, 1/25 IC50 level of E.P exhibited significant decline in GSH content (0.075 U/mg protein), followed by C.V (0.077 U/mg protein), and E.C (0.078 U/mg protein), respect to their control (0.087 U/mg protein) (Figure 6). However, 1/50 IC50 level of N.H exhibited significant decline in GSH content (0.071 U/mg protein), followed by E.P (0.076 U/mg protein), and E.C (0.081 U/mg protein). No significant differences were obtained between other toxins (Figure 6).
Genomic DNA diversity
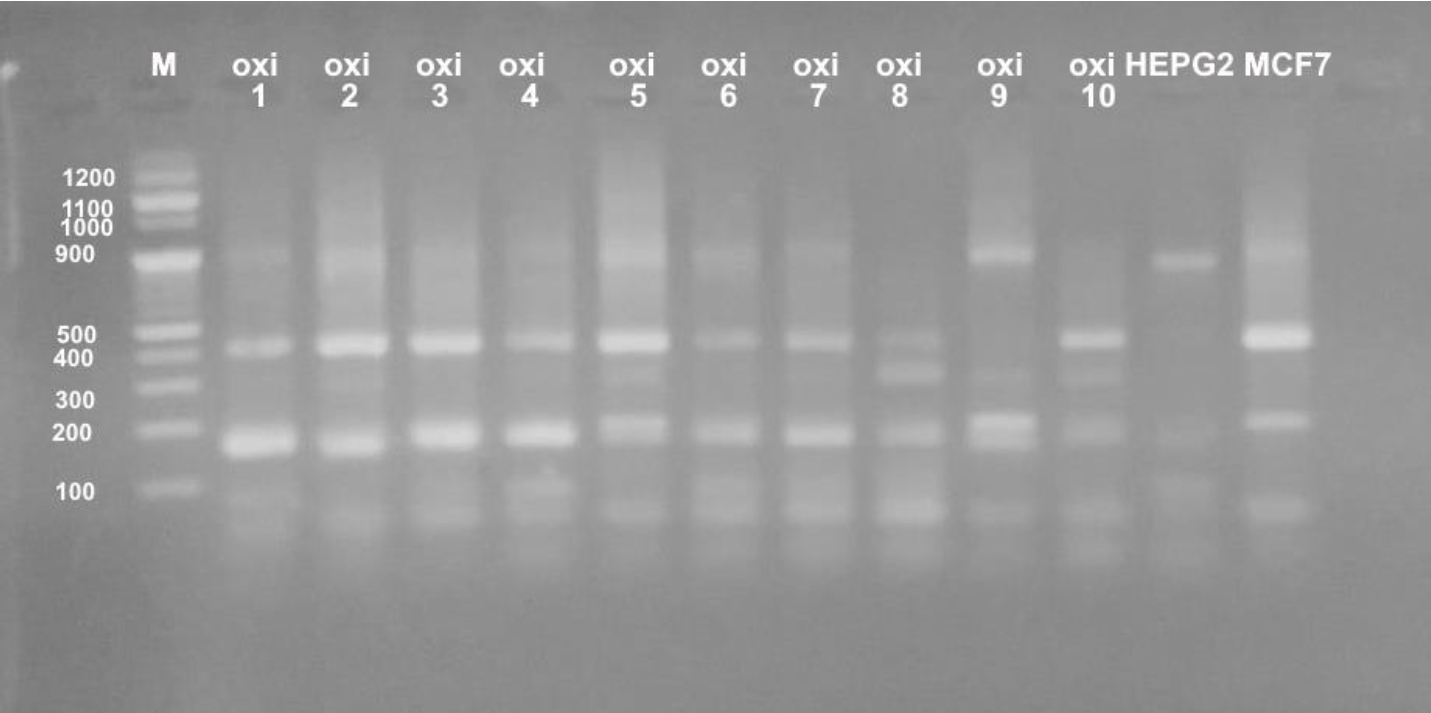
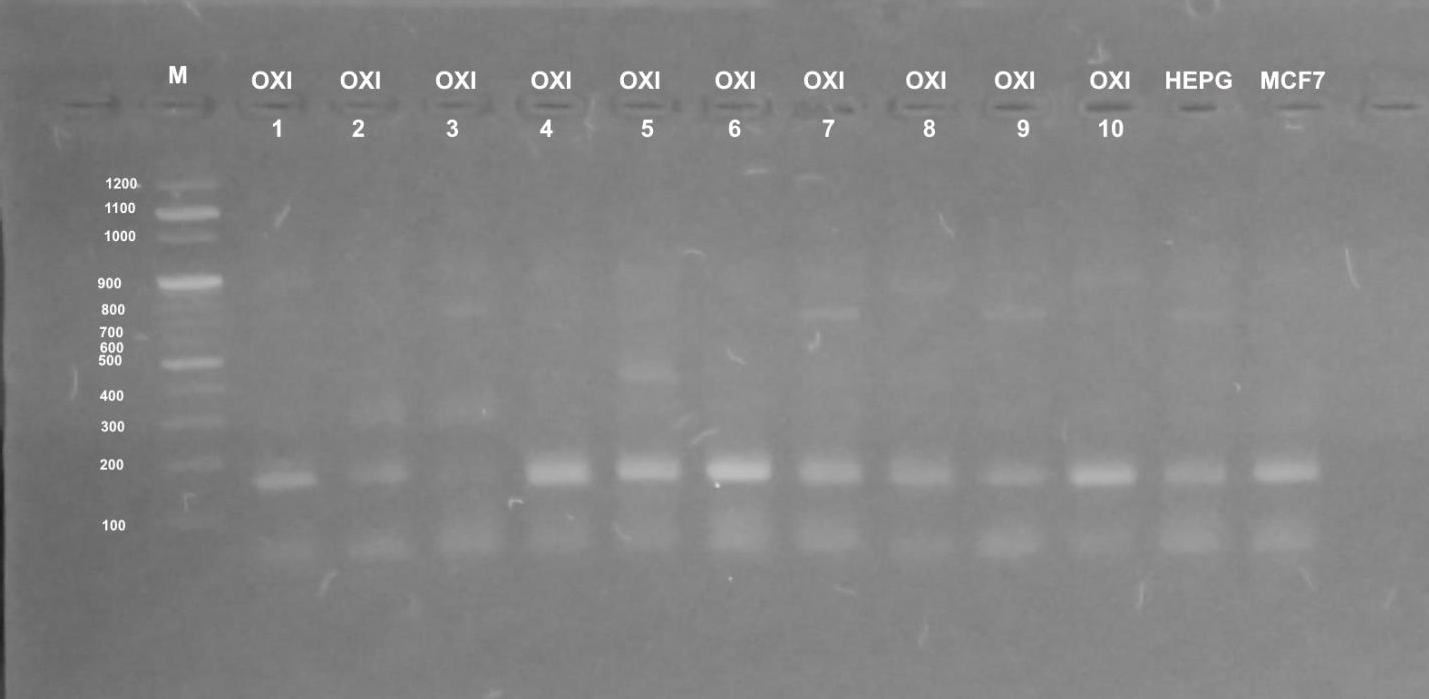
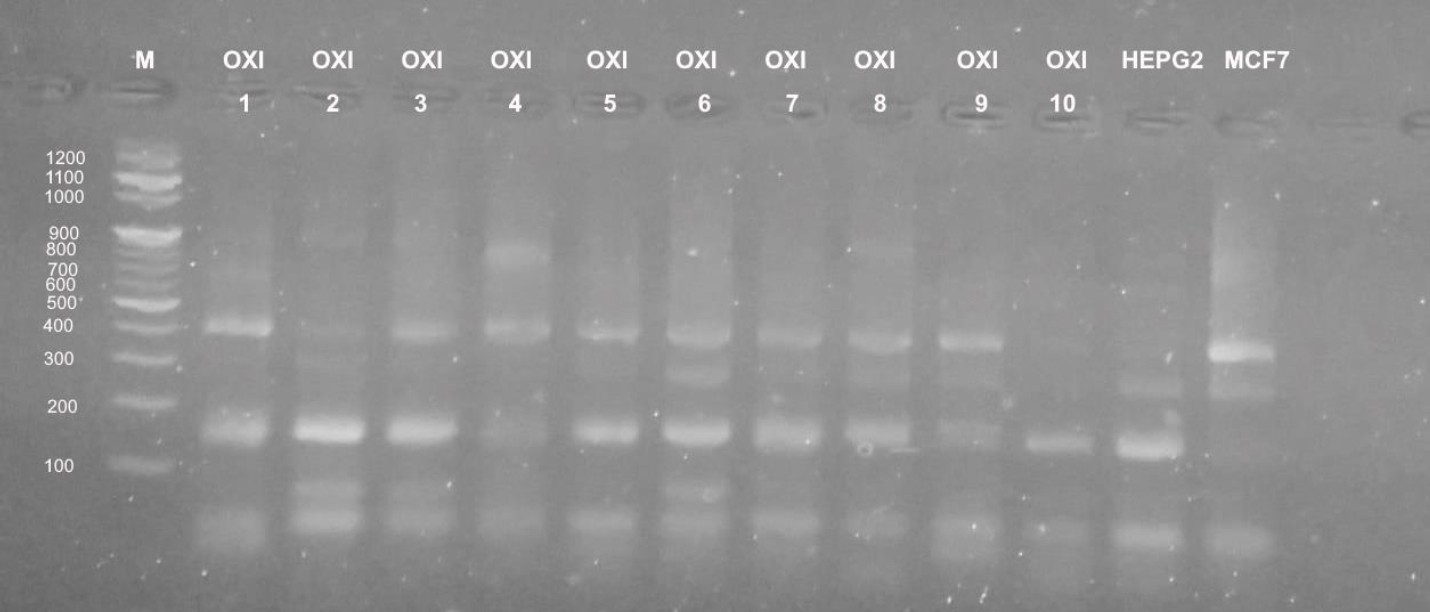
Different profiles were obtained when the genomic DNA of the used cultured mammalian cells that were exposed to doses of five Egyptian snake venom interacts (Figures 7-9). These profiles among genomic diversities of these cells were noticed after genomic template stability (GTS) estimates. There primers were employed to amplify genomic DNA successfully.
The RAPD fingerprints with varied bands ranged from 100 to 1000 b. p. were regulated by the state of treated and control cells and the used primer. In Figure 7, primer UBC-438 produced bands from 100 to 900 b. p. in the most samples. HepG2 cells significantly showed weak bands at amplified 100, 200 and 900 b. p. with treatment of E.C (GTS, 2.0-folds). Significantly increased bands at 200, 400 and 900 b. p. were noted for E.P treatment (GTS, 2.0-folds). Similar pattern was noticed for N.H. and C.C. treatments (GTS, 2.0 and 2.5-folds). However, C.V. treatment increased the intensity of bands at 100, 200, 400 and 900 b. p.
In case of MCF-7 cells, significant intensity was noticed in bands at 100, 200, 500 b. p. GST% slightly was noticed at 900 b. p. Treatment of E.C venom showed intensity at 200 and 500 b. p. In case of E.P treatment, increased bands (n=4) was noticed (GTS, 1.25-folds). Other amplifications were cited for N.H. (n=6) (GTS, 2.25-folds) and C.V. (n=5) (GTS, 1.25-folds). Treatment of C.C. showed increased intensity in band of 200 b. p (GTS, 1.25-folds).
In case of primer OPB-08, the amplification was less than UBC-438. All amplifications obtained significant bands at 100 and 200 b. p, except treatments of E.P., N.H. and C.V. showed slight intensity at 300, 400 and 900 b. p. (Figure 8). UBC-283 primer produced 3 bands in HepG2 control samples (Figure 9). Significant intensity was produced in bands at 200 and 400 b. p in case of E.C. treatment. The all amplifications showed intensity bands at 100 b. p. Low intensity of bands was obtained in C.C. treatment, although it contained 4 bands at 100, 200, 400 and 900 b. p. In case of MCF-7, treatment E.C. showed greatest amplification (band; n=5), followed by N.H. and C.C. treatment (n=4). However C.V. treatment exhibited the least amplification (n=2).
Discussion
Research into venomous systems offers significant insights into the biological roles of venom proteins [28] and provides useful information that can be utilized for effectively treating human snakebite [29]. Also, this trend provides potential avenue for novel drug discovery and design [30, 31]. Venom composition varies between species depending on several factors, e.g. phylogenetic affinities [32], geographic localities [33, 34], snake age [35-38] and diet [39, 40].
These compounds may serve as drug leads to combat cancer and may have potential for cancer treatment [41]. In vivo and in vitro experiments, it was shown that the snake toxins have anticancer effects on tumor and may have applications in cancer therapy. They show antitumor activity by killing the tumor cells directly, inhibiting tumor angiogenesis, or suppressing tumor growth [42]. Toxins from cobra venom exert a killing action on tumor cells and are widely studied for their anticancer properties. In the present work, the examined venoms showed variation between two cell lines: HepG2 and MCF-7 among toxicity. Such this pattern is in accordance with that obtained by Takahashi and Suzuki (1993) [43], where venom from the colubrid Ahaetulla nasuta was non-toxic toward A-375 cells, but it was quite anticancer potent toward MCF-7 cells.
Differences in ontcogeny and phenotype between these cell lines may explain the discrepancy observed between them. Also, several differences have been documented. For example, mammary gland cells are derived from embryologic mesenchyme, while melanocytes originate from cells of the neural crest. MCF-7 breast cancer cells are known to produce insulin-like growth factors and are responsive to estradiol, where they express cytoplasmic estrogen receptors. Also, these cells express WNT7B oncogene [44].
The ability of some snake venom toxins to cause anticancer potency is associated with their high specificity and affinity for cell and tissues. In spite of their anticancer effects, several isolated snake venom proteins and peptides have practical applications as pharmaceutical agents [45]. Moreover, snake venoms are able to act upon tumor cells has been known for a long-time. The first reported studies on using snake venom against tumor cells were related to the defibrination process. It was suggested that Ancrod, a polypeptide from Agkistrodon rhodostoma, administered with cyclophosphamide, could produce defibrination, thus decreasing the tumor weight by fibrinolysis and contributing to both detachment and decreased spread of some tumors [46]. However, such these findings showed that, besides defibrination, other complex mechanisms including platelet aggregation could be involved in the process. Braganca et al. (1976) [47] and (1985) [48] assayed a small fraction of N.H venom on cell cultures of Yoshida sarcoma, calling it cobra venom factor (CVF). Kaneda et al. (1985) [49] studied the anti-tumoral potential of purified peptides (cardiotoxin and cytotoxin) from the N.H atra snake. Then, Chiam et al. (1987) [50] showed the anticancer properties of several crude venoms from terrestrial snakes with lytic effects on cultures of malignant melanoma tumor cells.
Anticancer effects of snake venoms, show variations between species as well as tested cell lines, which could be linked in previous studies to variable modes of actions and receptor interactions of the molecules, making them attractive sources for cancer therapy [14, 51-54]. As stated in the literature, crude venom of C. cerastes from North Africa and C. purpureomaculatus from Asia were screened for anticancer action by performing a MTT assay on eight different cancerous human cell lines together with one non-cancerous cell line. Compared to our results, previous studies on closer related members of the Viperidae family (Montivipera bulgardaghica, Montivipera raddei and Protobothrops flavoviridis) indicated lower cytotoxicity against comparable cell lines [54, 55]. Furthermore, the venom of the related Indian Russell's viper (V. russelli) was tested for anticancer effects in an animal model of carcinoma and resulted in increased lifespan of mice, as well as cytotoxicity on leukemic cells in vitro [56]. A study on C. cerastes venom treated MCF-7 cells demonstrated a lower IC50 value (1.5 μg/ml) compared to another study (25.4 μg/ml) [12]. Additionally, the anti-cancer effects of an isolated LAAO from the close related C. vipera venom against MCF-7 cells was reported with an IC50 value of 2.8 μg/ml [22].
The ability of C. cerastes and V. alebetina snake venoms to stimulate apoptosis in human breast cancer cells and the related modulation of genes. The researchers evaluated the anticancer effect of the snake venoms on breast cancer (MCF-7) cells and found that the venoms exhibited concentration and time-dependent cytotoxicity. Typical apoptotic morphological features in venom-treated cells were visualized [12]. Also, venom toxin from V. alebetinaturanica inducing apoptosis of colon cancer cells via up-regulation of ROS-and JNK-mediated death receptor expression in colon cancer cells [18, 15]
More investigations must be followed to obtain better therapeutic treatment. Thus, some researchers found that the irradiated venom showed higher anticancer effect than crude venom, while the combination of crude venom and propolis extract demonstrated the highest effects against lung and prostate cancer cells [23].
The present results are in accordance with that observed in the cytotoxicity of other compounds in vitro models. The increased MDA levels in the cells explain the success of LPO process and the failure of antioxidant defense mechanisms to prevent the formation of excessive free radicals [59]. Enzyme, LDH catalyzes the translation of pyruvate to lactate and is careful to be a solution checkpoint of anaerobic gycolysis. It is one of the cellular death biomarkers that can be measured in many cases, where its release in the media of cultured cells is associated with cell and membrane damage [57, 58]. It is prominent in a lot of types of cancers and has been associated with tumor growth, maintenance, and invasion; so its reserve may limit the energy deliver in tumors and thus decrease the metastatic and invasive potential of cancer cells. This enzyme is getting a great deal of consideration as a potential diagnostic marker or a predictive marker of many types of cancer and as a therapeutic target for new anticancer treatments [60]. In the present study, the coupling increase in MDA and LDH in cell lysate and media is considered a good biomarker for the cytotoxic effect of the examined toxins. Recently, the anticancer effects of the examined toxins on cells MCF-7 and HepG2 are associated with oxidative stress and cell damage induction, whereas, during apoptosis, the potential difference in mitochondrial membrane is lost and some proteins such as Cytochrome C are released into the cytosol. When mitochondria are damaged, the antioxidant system is compromised resulting in an increased production of ROS and high induction of oxidative stress induced dose-dependent cytotoxicity characterized by cell death attributing to apoptotic and necrosis mechanisms, alterations of oxidative stress enzymes activity, increased levels of LPO, and LDH activity are considered biomarkers concerning cytotoxicity of venom’s toxins displays true results in this issue without any contrast in this fact [60].
The data exhibited wide ranges of values and variability for the examined snake venoms from different locations of the Egyptian environment. Such variability may be the result of cell characters and genotypes. Low doses or concentrations of the toxins indicated high inhibition of cancer cells under in vitro assays. A significant source of uncertainty and/or limitations is the action of such toxins in vivo assessments, because the estimated absorbed doses and the absorption, distribution, bioavailability, and metabolism patterns of the toxins are reliable concepts may be discussed, before further investigation for therapeutic agents’ conduction and fabrication. Finally, the present findings indicated the susceptibility of MCF-7 than HepG2 cells for the examined venoms. Such this reason is independent to different factors. Future research on venom proteomics can justify such differentiating activity which can be attributed to variation in venom constituents.
Conclusions
The study focused on the anticancer pattern of five venom’s snake collected from different locations of the Egyptian environment. MTT test (in vitro assay) provides reliable tool to assess interaction of drug with the cellular components under optimizing conditions of the incubation. The potency of the toxins on MCF-7 was more efficient than HepG2. Some biomarker profiles recognized the actions of the toxins into the cells. Increasing profiles of lactate dehydrogenase (LDH) activity and levels of glutathione content (GSH) and malodialdhyde (MDA) as well as repairment of DNA indicated such these actions. Future research on venom proteomics can justify such differentiating activity which can be attributed to variation in venom constituents. Such these investigations may document biomolecules which induce anticancer action.